Most intriguing and most tantalising of all, is the fact that we humans still have virtually no idea how to interpret DNA in any meaningful way. It's only since 1953 that we've understood what DNA even is; and it's only since 2001 that we've been able to extract and to gaze upon instances of the complete human genome.
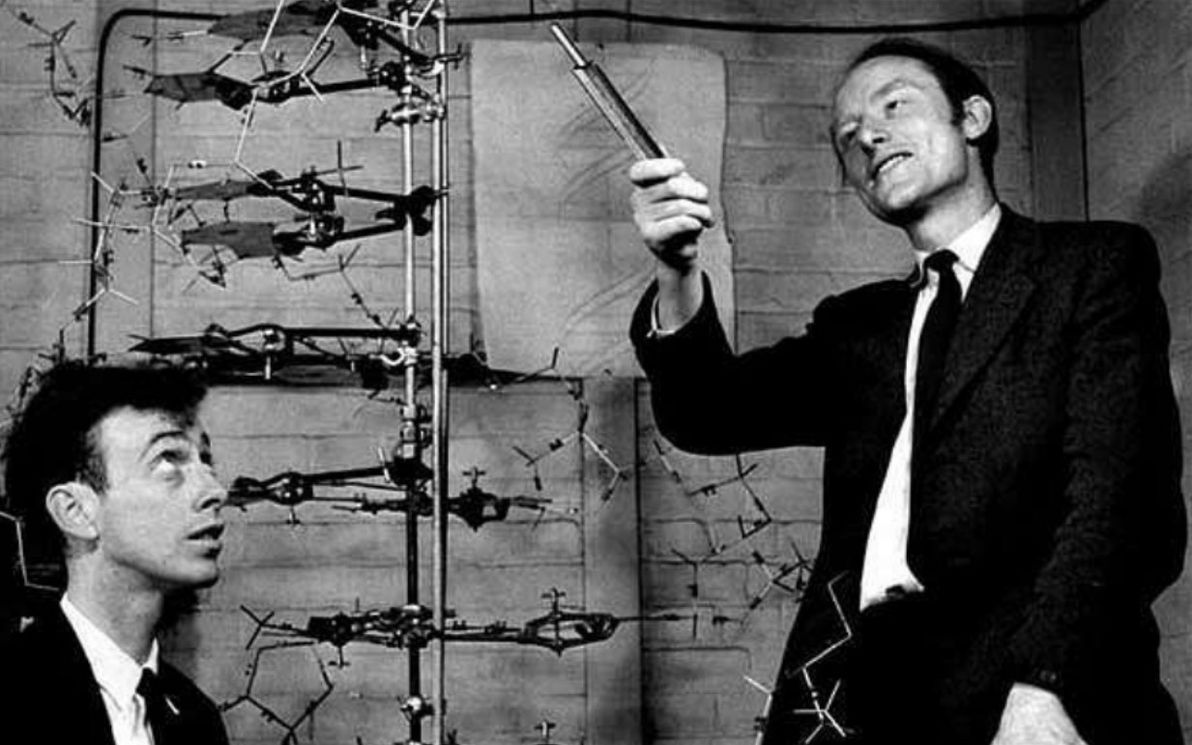
Image source: A complete PPT on DNA (Slideshare).
As others have pointed out, the reason why we haven't had much luck in reading DNA, is because (in computer science parlance) it's not high-level source code, it's machine code (or, to be more precise, it's bytecode). So, DNA, which is sequences of base-4 digits, grouped into (most commonly) 3-digit "words" (known as "codons"), is no more easily decipherable than binary, which is sequences of base-2 digits, grouped into (for example) 8-digit "words" (known as "bytes"). And as anyone who has ever read or written binary (in binary, octal, or hex form, however you want to skin that cat) can attest, it's hard!
In this musing, I'm going to compare genetic code and computer code. I am in no way qualified to write about this topic (particularly about the biology side), but it's fun, and I'm reckless, and this is my blog so for better or for worse nobody can stop me.
Authorship and motive
The first key difference that I'd like to point out between the two, is regarding who wrote each one, and why. For computer code, this is quite straightforward: a given computer program was written by one of your contemporary human peers (hopefully one who is still alive, as you can then ask him or her about anything that's hard to grok in the code), for some specific and obvious purpose – for example, to add two numbers together, or to move a chomping yellow pac-man around inside a maze, or to add somersaulting cats to an image.
For DNA, we don't know who, if anyone, wrote the first ever snippet of code – maybe it was G-d, maybe it was aliens from the Delta Quadrant, or maybe it was the random result of various chemicals bashing into each other within the primordial soup. And as for who wrote (and who continues to this day to write) all DNA after that, that too may well be The Almighty or The Borg, but the current theory of choice is that a given snippet of DNA basically keeps on re-writing itself, and that this auto-re-writing happens (as far as we can tell) in a pseudo-random fashion.

Image source: Art UK.
Nor do we know why DNA came about in the first place. From a philosophical / logical point of view, not having an answer to the "who" question, kind of makes it impossible to address the "why", by defintion. If it came into existence randomly, then it would logically follow that it wasn't created for any specific purpose, either. And as for why DNA re-writes itself in the way that it does: it would seem that DNA's, and therefore life's, main purpose, as far as the DNA itself is concerned, is simply to continue existing / surviving, as evidenced by the fact that DNA's self-modification results, on average, over the long-term, in it becoming ever more optimally adapted to its surrounding environment.
Management processes
For building and maintaining computer software, regardless of "methodology" (e.g. waterfall, scrum, extreme programming), the vast majority of the time there are a number of common non-dev processes in place. Apart from every geek's favourite bit, a.k.a. "coding", there is (to name a few): requirements gathering; spec writing; code review; testing / QA; version control; release management; staged deployment; and documentation. The whole point of these processes, is to ensure: that a given snippet of code achieves a clear business or technical outcome; that it works as intended (both in isolation, and when integrated into the larger system); that the change it introduces is clearly tracked and is well-communicated; and that the codebase stays maintainable.
For DNA, there is little or no parallel to most of the above processes. As far as we know, when DNA code is modified, there are no requirements defined, there is no spec, there is no review of the change, there is no staging environment, and there is no documentation. DNA seems to follow my former boss's preferred methodology: JFDI. New code is written, nobody knows what it's for, nobody knows how to use it. Oh well. Straight into production it goes.
However, there is one process that DNA demonstrates in abundance: QA. Through a variety of mechanisms, the most important of which is repair enzymes, a given piece of DNA code is constantly checked for integrity errors, and these errors are generally repaired. Mutations (i.e. code changes) can occur during replication due to imperfect copying, or at any other time due to environmental factors. Depending on the genome (i.e. the species) in question, and depending on the gene in question, the level of enforcement of DNA integrity can vary, from "very relaxed" to "very strict". For example, bacteria experience far more mutation between generations than humans do. This is because some genomes consider themselves to still be in "beta", and are quite open to potentially dangerous experimentation, while other genomes consider themselves "mature", and so prefer less change and greater stability. Thus a balance is achieved between preservation of genes, and evolution.
The coding process
For computer software, the actual process of coding is relatively structured and rational. The programmer refers to the spec – which could be anything from a one-sentence verbal instruction bandied over the water-cooler, to a 50-page PDF (preferably it's something in between those two extremes) – before putting hands to keyboard, and also regularly while coding.
The programmer visualises the rough overall code change involved (or the rough overall components of a new codebase), and starts writing. He or she will generally switch between top-down (focusing on the framework and on "glue code") and bottom-up (focusing on individual functions) several times. The code will generally be refined, in response to feedback during code review, to fixing defects in the change, and to the programmer's constant critiquing of his or her own work. Finally, the code will be "done" – although inevitably it will need to be modified in future, in response to new requirements, at which point it's time to rinse and repeat all of the above.
For DNA, on the other hand, the process of coding appears (unless we're missing something?) to be akin to letting a dog randomly roll around on the keyboard while the editor window is open, then cleaning up the worst of the damage, then seeing if anything interesting was produced. Not the most scientific of methods, you might say? But hey, that's science! And it would seem that, amazingly, if you do that on a massively distributed enough scale, over a long enough period of time, you get intelligent life.

Image source: DogsToday.
When you think about it, that approach isn't really dissimilar to the current state-of-the-art in machine learning. Getting anything approaching significant or accurate results with machine learning models, has only been possible quite recently, thanks to the availability of massive data sets, and of massive hardware platforms – and even when you let a ML algorithm loose in that environment for a decent period of time, it produces results that contain a lot of noise. So maybe we are indeed onto something with our current approach to ML, although I don't think we're quite onto the generation of truly intelligent software just yet.
Grokking it
Most computer code that has been written by humans for the past 40 years or so, has been high-level source code (i.e. "C and up"). It's written primarily to express business logic, rather than to tell the Von Neumann machine (a.k.a. the computer hardware) exactly what to do. It's up to the compiler / interpreter, to translate that "call function abc" / "divide variable pqr by 50" / "write the string I feel like a Tooheys to file xyz" code, into "load value of register 123" / "put that value in register 456" / "send value to bus 789" code, which in turn actually gets represented in memory as 0s and 1s.
This is great for us humans, because – assuming we can get our hands on the high-level source code – we can quite easily grok the purpose of a given piece of code, without having to examine the gory details of what the computer physically does, step-by-tiny-tedious-step, in order to achieve that noble purpose.
DNA, as I said earlier, is not high-level source code, it's machine code / bytecode (more likely the latter, in which case the actual machine code of living organisms is the proteins, and other things, that DNA / RNA gets "compiled" to). And it now seems pretty clear that there is no higher source code – DNA, which consists of long sequences of Gs, As, Cs, and Ts, is the actual source. The code did not start in a form where a given gene is expressed logically / procedurally – a form from which it could be translated down to base pairs. The start and the end state of the code is as base pairs.

Image source: The University Network.
It also seems that DNA is harder to understand than machine / assembly code for a computer, because an organic cell is a much more complex piece of hardware than a Von Neumann-based computer (which itself is a specific type of Turing machine). That's why humans were perfectly capable of programming computers using only machine / assembly code to begin with, and why some specialised programmers continue primarily coding at that level to this day. For a computer, the machine itself only consists of a few simple components, and the instruction set is relatively small and unambiguous. For an organic cell, the physical machinery is far more complex (and whether a DNA-containing cell is a Turing machine is itself currently an open research question), and the instruction set is riddled with ambiguous, context-specific meanings.
Since all we have is the DNA bytecode, all current efforts to "decode DNA" focus on comparing long strings of raw base pairs with each other, across different genes / chromosomes / genomes. This is akin to trying to understand what software does by lining up long strings of compiled hex digits for different binaries side-by-side, and spotting sequences that are kind-of similar. So, no offense intended, but the current state-of-the-art in "DNA decoding" strikes me as incredibly primitive, cumbersome, and futile. It's a miracle that we've made any progress at all with this approach, and it's only thanks to some highly intelligent people employing their best mathematical pattern analysis techniques, that we have indeed gotten anywhere.
Where to from here?
Personally, I feel that we're only really going to "crack" the DNA puzzle, if we're able to reverse-engineer raw DNA sequences into some sort of higher-level code. And, considering that reverse-engineering raw binary into a higher-level programming language (such as C) is a very difficult endeavour, and that doing the same for DNA is bound to be even harder, I think we have our work cut out for us.
My interest in the DNA puzzle was first piqued, when I heard a talk at PyCon AU 2016: Big data biology for pythonistas: getting in on the genomics revolution, presented by Darya Vanichkina. In this presentation, DNA was presented as a riddle that more programmers can and should try to help solve. Since then, I've thought about the riddle now and then, and I have occasionally read some of the plethora of available online material about DNA and genome sequencing.
DNA is an amazing thing: for approximately 4 billion years, it has been spreading itself across our planet, modifying itself in bizarre and colourful ways, and ultimately evolving (according to the laws of natural selection) to become the codebase that defines the behaviour of primitive lifeforms such as humans (and even intelligent lifeforms such as dolphins!).

Image source: Days of the Year.
So, let's be realistic here: it took DNA that long to reach its current form; we'll be doing well if we can start to understand it properly within the next 1,000 years, if we can manage it at all before the humble blip on Earth's timeline that is human civilisation fades altogether.
]]>